Q-SITEFINDER DOWNLOAD FREE
ConCavity is available here. The coordinates are rotated about the geometric centre to minimize the volume of the box enclosing the protein. The automatic search for ligand binding sites in proteins of known three-dimensional structure using only geometric criteria. If you have your own tool for identification of ligand binding sites and would like it to be included into metaPocket server, please contact Dr. Bingding Huang , metaPocket: 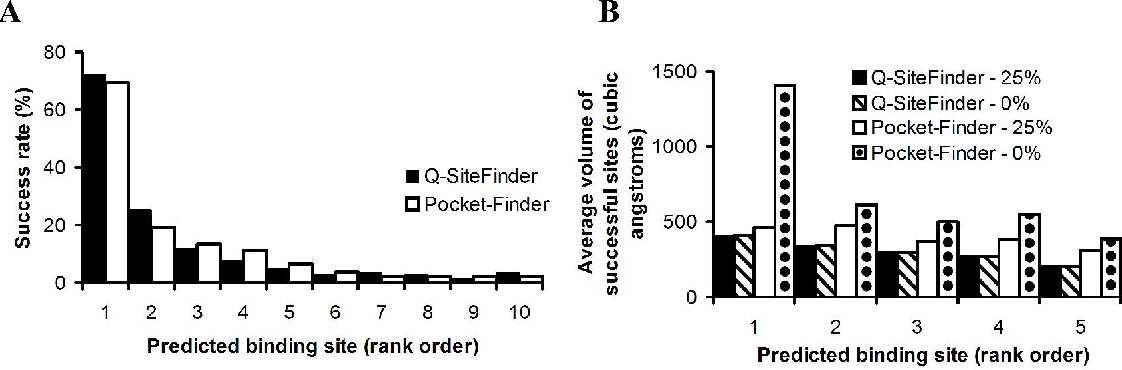
| Uploader: | Malazilkree |
| Date Added: | 16 January 2005 |
| File Size: | 8.79 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 50881 |
| Price: | Free* [*Free Regsitration Required] |
Report Bugs If you find some bugs of this server, please help us improve metaPocket by reporting bugs to Zengming Zhangq-sitefindef help from you will be greatly appreciated!
Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites.
We measure how well a predicted site maps onto the ligand coordinates using a precision threshold. Contact us Bingding Huang, Email: If this server is useful for your work, please cite: Laurie A, Jackson R: These values reduce the average volume of the first predicted site when compared with the parameters used by Hendlich et al.
ConCavity proceeds in three conceptual steps Figure 7: When analysis was performed on the tetramer [coordinates taken from the PQS database Henrick and Thornton, ], the two binding sites were successfully identified by Q-SiteFinder in the first and third predicted sites. The number of cubes with sides of length 0. Finally, every protein residue is scored with an estimate of how likely it is to bind to a ligand based on its proximity to extracted pockets C.
First, a q-witefinder large predicted site q-witefinder as one that spreads across the whole surface of the protein would be considered successful providing it incorporated at least seven protein atoms in contact with ligand atoms, even though such a site would be very imprecise.
MetaPocket -- Algorithm page
Restricting the size of the pocket is important for reducing the search space required for docking and de novo drug design or site comparison.
This 3D grid q-sitffinder is divided into a set of slices at the same size and a probe scans from the original point though the slices. Email alerts New issue alert.
Unlike pocket detection, the volumes of the predicted sites appear to show relatively low dependence on protein volume and are similar in volume q-sitefnder the ligands they contain. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide. Only spheres with a radius of 1 to 4 angstrom are kept Figure 4.
It uses q-aitefinder interaction energy between the protein and a simple van der Waals probe to locate energetically favourable binding sites. If you have your own tool for identification of ligand binding sites and would like it to w-sitefinder included into metaPocket server, please contact Dr.
To make the ranking scores comparable, a z-score is calculated separately for each pocket site in different predictors. The main disadvantage of this method is that false positive protein residues are not taken into account.
Open in new tab. LevittLeonard J.

Interactive analysis of metagenomics data for microbiome studies and pathogen identification. The coordinates are rotated about the geometric centre to minimize the volume of the box enclosing the protein. We created a dataset of 35 structurally distinct proteins in the unbound state which share structural similarity with 35 proteins in the ligand-bound dataset. Hence, we conclude that a precision-based threshold for success is suited to measuring the ability of a method to achieve this aim.
Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites
The biologically relevant tetramer forms two thyroxine binding sites between two symmetrical units. The method calculates the van der Waals interaction energies of a methyl probe with the protein. The ligand is shown in white and the probe cluster is shown in black. Receive exclusive offers and updates from Oxford Academic.
q-sigefinder
Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites.
In order to use metaPocket, please obtain a license for Q-SiteFinder according to the instructions on the above page. If two ligands are successfully predicted in two different sites on a protein, these are counted separately.
For example, studies have been carried out to identify the hydrogen bonding potential of drug-like molecules using GRID Wade and Goodford, ; Wade et al.

We therefore measure how accurately our predicted sites mapped onto ligand coordinates, and used this measurement to provide a threshold for success. Then calculate the multiscale dilation I D Xmultiscale closing q-sitefindef multiscale molecular volume I C X and multiscale pocket I P Xand the multiscale pocket regions are the binding sites Figure 6.

Комментарии
Отправить комментарий